Analysis
The analysis interface offers two basic ways to select proteins for analysis. 1) A user enter select a valid PDB ID. If the this entry has already been analyzed, the results will be presented, otherwise the server will automatically retrieve the needed sequence data from the PDB and begin processing it. 2) A user may submit custom data in FASTA format. The user will also be required to enter a 4-letter alphanumeric code to identify the submission and will then be provided with a link to the output for the sequence.
Figure 1 is a snapshot from the page and shows the options available to the user. The first part of the form allows the user to select the source of data to use while the second part requires the user to enter an email address so they can be notified should analysis have needed to be performed.
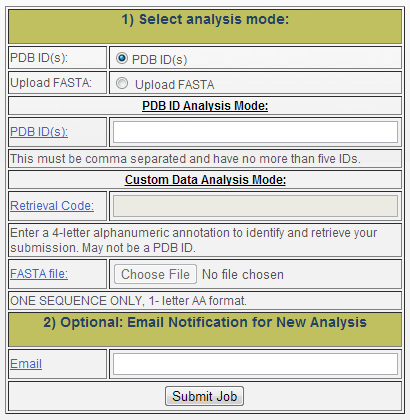
Figure 1: MIR analysis form
- "PDB ID(s)": Here the user may enter Protein Data Bank (PDB) [1] IDs for analysis. Up to five PDB IDs may be entered here, provided that they are separated by commas. The follow page will either provides links to the existing analysis or provide links that will be populated once automatic analysis has completed. Note that proteins longer than 500 residues cannot be run automatically. If you are interested in running longer proteins, please contact us.
- "Retrieval Code": Initially, the user must provide a retrieval that will be used to track the protein in our system. This must be a four letter alphanumeric string that is not a valid PDB ID. Hence, the user must remember the code they have selected in order to retrieve its data or save the link provided on the following page.
- "FASTA file": The user must browse and select a text FASTA file from their local computer. The FASTA file must contain exactly one sequence and contain the protein in the 1-letter AA format.
- "Email:" Optionally the user may enter an email to receive a notification in the case that analysis results are not already available. The address will not be used for any other process.
References
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N. and Bourne, P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235-242.
- Papandreou, N., Berezovsky, I.N., Lopes, A.,
Eliopoulos, E. and Chomilier, J. (2004) Universal positions in
globular proteins. Eur. J. Biochem., 271, 4762-4768.
- Chomilier, J., Lamarine, M., Mornon, J.-P.,
Torres, J.H., Eliopoulos, E. and Papandreou, N. (2004) Analysis of
fragments induced by simulated lattice protein folding. C R Biol.,
327, 431-443.
- Bostock, M., Ogievetsky, V., and Heer, J. (2011) D3: Data-Drive Documents. IEEE Trans. Visualization & Comp. Graphics (Proc. InfoVis) .